Source: venturebeat.com
Google’s latest research dives deep into the brains of fruit flies — quite literally. In collaboration with the Howard Hughes Medical Institute (HHMI) Janelia Research Campus and Cambridge University, the tech giant today published the results of a study (“Automated Reconstruction of a Serial-Section EM Drosophila Brain with Flood-Filling Networks and Local Realignment“) that explores the automated reconstruction of an entire fly’s brian neuron by neuron.
It’s a spiritual follow-up to a paper published in the journal Cell by researchers at Janelia Research Campus. The team involved in that study infused a fly brain’s cells and synapses with heavy metals to mark the outlines of each neuron and its connections. To generate images, they hit approximately 7,062 brain slices with a beam of electrons, which passed through everything except the metal-loaded parts.
The coauthors of this most recent paper expect that their work in connectomics — the production and study of connectomes, or comprehensive maps of connections within an organism’s nervous system — will accelerate investigations at HHMI and Cambridge University into learning, memory, and perception in the fly brain. In the spirit of open source, they’ve made the full results available for download and browsable online using Neuroglancer, an in-house interactive 3D interface.
lies in the genus drosophila weren’t an arbitrary target. As several of the paper’s coauthors note in an accompanying blog post, fly brains are relatively small (one hundred thousand neurons) compared to, say, frog brains (over 10 million neurons), mouse brains (100 million neurons), octopus brains (half a billion neurons), or human brains (100 billion neurons). That makes them easier to study “as a complete circuit,” they say.
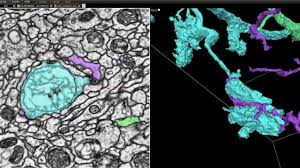
Plotting out a fly brain required first sectioning it into thousands of ultra-thin 40-nanometer slices, which were imaged using a transmission electron microscope and aligned into a 3D image volume of the entire brain. Next, thousands of Cloud tensor processing units (TPUs) — AI accelerator chips custom-designed by Google — ran a special class of algorithm called flood-filling networks (FFNs) designed for instance segmentation of complex and large shapes, particularly volume data sets of tissue. Over time, the FFNs automatically traced each individual neuron in the fly brain.
Reconstruction didn’t go off without a hitch; the FFNs performed poorly when image content in consecutive sections wasn’t stable or when multiple consecutive slices were missing (due to challenges associated with the sectioning and imaging process). To mitigate dips in precision and accuracy, the team estimated the slice-to-slice consistency in the 3D brain image and locally stabilized the content while the FFNs highlighted each neuron. Additionally, they used an AI model dubbed Segmentation-Enhanced CycleGAN (SECGAN) — a type of generative adversarial network specialized for segmentation — to computationally fill in missing slices in the image volume. With the two new procedures in place, they found that FFNs were able to trace through locations with multiple missing slices “much more robustly.”
With the brain fully imaged, the team tackled the problem of visualization with the aforementioned Neuroglancer, which is available in open source and currently in use by collaborators at the Allen Institute for Brain Science, Harvard University, HHMI, Max Planck Institute, MIT, Princeton University, and elsewhere. It’s based on WebGL and supported in newer versions of Chrome and Firefox, and it exposes a four-pane view consisting of three orthogonal cross-sectional views as well as a view (with independent orientation) that displays 3D models for selected objects.
In addition to enabling the viewing of petabyte-scale 3D volumes, Nueroglancer supports features like arbitrary-axis cross-sectional reslicing, line-segment-based models, multi-resolution meshes, and the ability to develop custom analysis workflows via integration with Python. Moreover, it’s able to ingest data via HTTP in a range of formats including BOSS, DVID, Render, precomputed chunk and mesh fragments, single NIfTI files, Python in-memory volumes, and N5.
The paper’s coauthors note that their brain image isn’t perfect, because it still contains some errors and skips over the identification of synapses. But they expect that advances in segmentation methodology will yield further improvements in reconstruction, and they say that they’re working with Janelia Research Campus’ FlyEM team to create a “highly verified” and “exhaustive” fly brain connectome using images acquired with focused ion beam scanning electron microscopy technology.